Tracking the evolution of cancer cell populations through the mathematical lens of phenotype-structured equations
Résumé
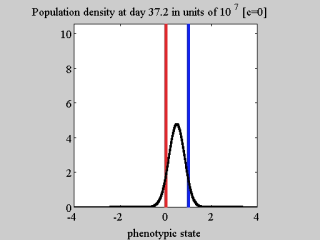
AbstractBackgroundA thorough understanding of the ecological and evolutionary mechanisms that drive the phenotypic evolution of neoplastic cells is a timely and key challenge for the cancer research community. In this respect, mathematical modelling can complement experimental cancer research by offering alternative means of understanding the results of in vitro and in vivo experiments, and by allowing for a quick and easy exploration of a variety of biological scenarios through in silico studies.ResultsTo elucidate the roles of phenotypic plasticity and selection pressures in tumour relapse, we present here a phenotype-structured model of evolutionary dynamics in a cancer cell population which is exposed to the action of a cytotoxic drug. The analytical tractability of our model allows us to investigate how the phenotype distribution, the level of phenotypic heterogeneity, and the size of the cell population are shaped by the strength of natural selection, the rate of random epimutations, the intensity of the competition for limited resources between cells, and the drug dose in use.ConclusionsOur analytical results clarify the conditions for the successful adaptation of cancer cells faced with environmental changes. Furthermore, the results of our analyses demonstrate that the same cell population exposed to different concentrations of the same cytotoxic drug can take different evolutionary trajectories, which culminate in the selection of phenotypic variants characterised by different levels of drug tolerance. This suggests that the response of cancer cells to cytotoxic agents is more complex than a simple binary outcome, i.e., extinction of sensitive cells and selection of highly resistant cells. Also, our mathematical results formalise the idea that the use of cytotoxic agents at high doses can act as a double-edged sword by promoting the outgrowth of drug resistant cellular clones. Overall, our theoretical work offers a formal basis for the development of anti-cancer therapeutic protocols that go beyond the ‘maximum-tolerated-dose paradigm’, as they may be more effective than traditional protocols at keeping the size of cancer cell populations under control while avoiding the expansion of drug tolerant clones.ReviewersThis article was reviewed by Angela Pisco, Sébastien Benzekry and Heiko Enderling.
Fichier principal
 13062_2016_Article_143.pdf (1.82 Mo)
Télécharger le fichier
13062_2016_143_MOESM1_ESM.mov (203.07 Ko)
Télécharger le fichier
13062_2016_Article_143.pdf (1.82 Mo)
Télécharger le fichier
13062_2016_143_MOESM1_ESM.mov (203.07 Ko)
Télécharger le fichier
 13062_2016_143_MOESM2_ESM.mov (415.28 Ko)
Télécharger le fichier
13062_2016_143_MOESM2_ESM.mov (415.28 Ko)
Télécharger le fichier
| Origine | Publication financée par une institution |
|---|
| Origine | Publication financée par une institution |
|---|
| Origine | Publication financée par une institution |
|---|
Loading...

